Lessons Learned from two years as a Data Scientist
What I learned from my first two years as a data scientist
Intro
I finally escaped from (grad) school in 2019, spent two months interning as an assistant trader at FTX, and have since spent the last two years working as a data scientist, with the bulk of that time at Microsoft and the last two months at Anthropic. Not gonna lie — my time as a data scientist has been pretty awesome. I was technically on a product team at Microsoft in DevDiv working on improving developer tools like VS Code, but in practice it was like a flush research job with tons of freedom (to first approximation my instructions for the first six months were, "Fix bugs using machine learning. Go.") I got to work with cutting edge transformer models and their application to source code, the most famous of which is autocompletion ala intellisense, tabnine, and Codex. Every two months the amount of compute I have access to has doubled, starting out with a 2013 macbook air and ending with a cluster with thousands of A100s (which, to be fair, I share with more people than my old laptop), and I got to collaborate on ten different papers.
I was extremely green when I started out. I hadn't used python since cs1 as a college freshman, excluding one month where I worked through fast.ai and leetcode. Maybe the most telling anecdote is that I spent an embarrassingly long time using vim as my exclusive file editor... while not knowing any of the vim shortcuts besides :wq (write and quit).

Since then I've become much more competent, in python, on the command line, as a member of a software engineering team, and as a machine learning researcher, and I wanted to share some of the things that I've learned along the way.
Python
Python is the language of data science. This is mostly because Python has fantastic libraries like numpy, pandas, and torch, as well as great tools like jupyter/colab and pdb (the python debugger), but also because it's a concise language that allows for quickly writing powerful scripts. There's no Java awfulness like
import java.util.ArrayList;
ArrayList<String> cars = new ArrayList<String>();instead it's just cars = [].
Notebooks
One of the most exciting tools for python to learn about is notebooks, especially Jupyter and colab. Notebooks are a type of repl (read-eval-print loop, see https://replit.com/ for repl's in many different languages), which allow for extremely fast iterative development. An old school development process typically involves rerunning an entire file over and over again each time you make a change. A notebook or repl however maintains state, so you can define the input variables once and then simply execute the lines that you're truly interested in.
Notebooks also enable some amazing tutorials and documentation, since you can interleave markdown, graphs, and executable code snippets in a notebook. It's very easy to imagine rewriting this blog post as a notebook :p. In practice my own notebooks tend to be very messy playgrounds, which I'll occasionally clean up to share with people.
Colab in particular is notable for granting (somewhat spotty) access to a free GPU from Google. Quite the service for the ML community! I find that colab notebooks are also easier to share than jupyter notebooks, although in general I like the jupyter UI more.
To get started with jupyter, you can go to the command line and do:
pip install jupyter-notebook
# often there will be an obnoxious step here
# where you need to add the jupyter binary path to your PATH variable, sorry
jupyter notebook # and follow the local linkOne trick for jupyter is to autoreload modules that you've edited. That way, say you have a notebook cell my_var = MyClass('hi') and change my_class.py, then MyClass will automatically change, and you won't have to reimport it (you will still have to reinitialize my_var).
%load_ext autoreload
%autoreload 2Another trick is that you can use ?? to see a function's definition, e.g. in jupyter:
from tqdm import tqdm
??my_fun(In general you can use the inspect library to retrieve definitions, without needing jupyter.) E.g.:
import inspect
def my_fun():
print(5)
inspect.getsource(my_fun)One of my favorite ways to get a handle on a new library is to import it into jupyter, use dir(that_lib) to see all the possible options, and then trying to run all of the ones which seem interesting.

Debugging
I used to think that coding was 90% debugging and only 10% actually writing the bulk of a project. I think there are a couple reasons for this. First because I was coming from a math background, where I felt like the hard work should be understanding the problem, and that actually writing the code should be as simple as writing down the solution to a problem after you've solved it. Second because I was a noob and would get stuck on frustratingly simple things as well as common gotchas for a long time. Third because I'm pretty sure I would attempt to write the entire program in one fell swoop and then painfully spend the next hour patching it up, rather than add functionality more incrementally like you do in a notebook. Finally, because I didn't know about the python debugger, pdb.
Say you have some buggy code like so:
def bad_fun():
my_list = ['1','0','2']
my_list = sorted(my_list)
assert my_list == ['0','1','2']
return my_list
bad_fun()Sure, you could add a print statement to the assert, like so:
assert my_list == ['0','1','2'], my_list. But to really see what's going on, you can insert a breakpoint import pdb; pdb.set_trace(), say after the line where my_list is defined. The program will run until your breakpoint, and then give you control. You can then inspect everything, e.g. p my_list, p sorted(my_list) etc. You can also execute arbitrary code, such as defining new variables like other_list = sorted(my_list). You can also use the commands u and d (for up and down) to navigate the call stack, which in this case would just have two frames: the highest-level frame which call bad_fun, and the deeper frame which actually sets my_list. Finally, you can continue executing the program you've interrupted using the commands for next (execute until the next line in the file), step (execute the next line in the program (which might mean going deeper into thecall stack), and continue (run the whole program, at least up until the next breakpoint).
pdb plays poorly with multiprocessing (by default import pdb; pdb.set_trace() will simply crash your program if its inside a child process you've spawned using multiprocessing.pool.Pool, so I always implement my programs with a single worker first before parallelizing.
One large class of buggy or otherwise subpar programs is code that is too slow. I almost always have some lightweight profiling going in the form of a progress bar like tqdm in order to track how long my data processing jobs will take.
from tqdm import tqdm
for i in tqdm(range(10)):
print(i)I feel really dumb when I start a job without tqdm, wait a minute, and then realize it could take anywhere between another minute and years to finish.

For more rigorous profiling, the standard is to use cprofile e.g.
python -m cprofile -o my_file_profiling.o my_file.py
and then in jupyter:
import pstats
from pstats import SortKey
p = pstats.Stats('my_file_profiling.o')
p.sort_stats(SortKey.CUMULATIVE).print_stats(10)It almost always makes sense to profile before doing any laborious optimization.
People at Anthropic especially like py-spy, because it can be used on a program that is already running, unlike cprofile.
Testing
Somewhat embarrassingly, I ran about one million tests written by other people as part of my DeepDebug project before writing my first test to test my own code. I think that testing is more important when other people are going to be editing your code (and potentially breaking it!), in part because it gives them a foothold to see how your code is run. At Anthropic we forbid merging to master unless all tests found by pytest pass.
Type Hints
Python does have a handful of shortcomings. Duck-typing, i.e. just doing cars = [], can be very confusing when working on a larger project, especially with collaborators. Since python's primitive objects are so powerful, it can be pretty tempting to just carry around a bunch of lists and dictionaries rather than define well-scoped classes for them. Note that this will also confuse the IDE, which won't be able to provide good goto or autocompletion support. The fix for this is pretty great, though. The first rule is to use type hints, like from typing import List; cars: List[str] = []. (In general these type hints are merely suggestions though, and won't actually be enforced.) The second tactic is to simply use classes more.

Duck-typing can also make a python project unsafe, and in fact Google doesn't allow any production-level projects to be written in python due to safety concerns. I hear on the street that especially rust is a safety-first language which is all the rage these days.
Parallelism
In theory Python can be a slow language, again due to duck-typing and being an interpreted language rather than a compiled one. In practice however, the heavy-lifting for ml is done by numpy and C code, and apps like webscraping are typically io-bound anyway.
Python has the notorious global-interpreter lock (GIL), which in theory makes parallelism more difficult. To be honest, I'm not sure how much truth there is to that. Debugging parallelized python programs is hard since you can't simply use import pdb; pdb.set_trace() the way you normally would, but is there any language in which debugging parallel programs is easy? In practice there are several good workarounds.

You can bypass the GIL by launching subprocesses e.g.
from multiprocessing.pool import Pool
with Pool() as pool:
res = pool.map(my_function,my_data)The standard library's multiprocessing.pool.Pool is problematic in that it has to copy state to each subprocess (a big deal if you have already loaded a massive data object into memory rather than streaming it in chunks or passing metadata to the pool workers!) A near drop-in replacement that solves this problem is ray's multiprocessing.
You can continue to use pdb for most of the development cycle if you first build a single-threaded version of your program before parallelizing.
Python also has this trick:
import traceback
def bad_fun():
try:
assert False
except:
return traceback.format_exc()which lets you grab stack traces from any worker that runs into an error and throws an exception.
And finally python has great support for spark, which makes it very easy to do things like keep running a data processing job in face of crashing workers, or to scale the number of cores up or down mid-run.
Imports
My least favorite part of python is the import system. You can see where python looks for projects that you're trying to import by doing:
import sys
print(sys.path)This should give a list of paths like
['/Users/dawndrain/Code/blog_post_dir',
'/opt/anaconda3/lib/python3.7',
'/Users/dawndrain/.local/lib/python3.7/site-packages',
]Note that the first path is your current working directory. In this example the second path points to the standard python library functions, and the third path points towards any packages that you've installed with pip. If your program does something like from my_package import my_function, then python will look along those paths in order until it finds my_package, and then try and import my_function from it. For the longest time I did not know how to import my own code across projects! I had this one dawn_utils.py file which I must have copy-pasted like five times. The solution is to add your project's folder to one of the directories on the PYTHONPATH (what sys.path shows). In practice the way to do this is to go to your project's root folder (which should contain a subfolder like project_src that has your project's actual code) and create a setup.py file like:
from setuptools import setup
setup(
name='my-project',
version='0.0',
packages=['project_src',],
install_requires=[],
long_description=open('readme.txt').read(),
)You can then do pip install . or pip install -e . (which will make it so that you can edit your project and have the changes propagate without having to reinstall it). You should now be able to do import project_src and go to town :).
Env Management
One of the first words that comes to mind when I think of environment management is "dependency hell". Strictly speaking, dependency hell is when the packages your project depends on have their own conflicting dependencies e.g. package A requires version 1 of a library, and package B requires version 2. In pathos, dependency hell is any time I feel like I'm spending more time just setting up dependencies (pypi packages, teammates' code, github repos, sometimes awful stuff like the correct CUDA version, macOS vs. linux vs. windows etc.) than I am actually writing code.

There are a few ways to make environment management less painful. You can create a clean python environment by doing python -m venv my_venv && source my_venv/bin/activate. (You can also use conda env's instead of venv). This will create a local site-packages folder, which is where pip will install new packages so long as the venv is active. If you want to share your list of installed packages with someone else, it's as easy as doing pip freeze > requirements.txt, which someone can then reinstall using pip install -r requirements.txt. You can also programmatically add those reqs to your setup.py by adding install_requires = open('requirements.txt','r').readlines().
Note that this is specific to python, and doesn't work for OS packages like tree or ncdu which are installed with apt-get (linux) or homebrew (mac). In that case you'll need to use a docker image, which thankfully can be shared across teammates.
For working with teammates, GitHub (or a competitor like Azure Devops) is essential. We use a monorepo at Anthropic, similar to all of Google(!), which makes it much easier for everyone's environment to stay in sync. It also makes it much easier to use other people's code and prevent needless duplication.
Books
As a rule, reading programming books is a waste of time (the notable exception being delightful mathy books like CLRS's Introduction to Algorithms or Skiena's The Algorithm Design Manual, which are written in pseudocode and not so much about programming per se). If you've just read three pages in a row about some programming language without having written any code in that language in the meantime, you are probably doing something wrong. That being said, I did like reading Dan Bader's Python Tricks the Book this one evening, which taught me about list comprehensions, generators, *args and **kwargs, context managers, env management and lots of other goodies. Honorable mentions if you have to read another book about python include The Python Cookbook, The Hitchhiker's Guide to Python, and Fluent Python (very long).

The Command Line
You can access the command line using an app like terminal, or gitbash or iterm.
You navigate the command line using cd (change directory), can see what files are there using ls (shows file in the current directory), tree (recursively shows files), ncdu (shows files along with sizes), and find. You can read the files using cat, head and tail (like cat but just the beginning or end), less (kind of like an interactive cat that you can navigate using vim commands), jq (specially for reading json files). You read just the parts of a file that satisfy a regex by doing cat my_file.txt | grep my_regex (the vertical line is pronounced 'pipe' and lets you chain together commands.) You can copy files using cp and its big brother rsync (which is very useful for updating data across a network, as it both automatically compresses and only sends the diff), move files with mv, delete them with rm and troll your friends with rm -rf (force-deletes everything recursively!!). You can see how your storage is being used with du or df -h, and see how much memory is being used with free -h. You can check on what processes are currently running with (h)top or ps -axf. watch is useful for continuously running a command, e.g. watch -n1 nvidia-smi will show you the output of nvidia-smi every second. You can edit files with vim if you must. Once in vim, the most important command is :q. You can see your command history using history (history | grep cmd is pretty helpful). One of the most important commands is ssh, which allows you to control a (typically more powerful) computer remotely from your laptop. You can also forward ports so that you can locally see the remote machine's localhost/6006 or whatever.
Arguably the most important command is python ;), which calls a python script or, if used without an argument, starts a python session (you can also use ipython, which is slightly better for interactive command line sessions), there's also the command bash for executing shell scripts.
You can run run processes in parallel using cmd1 & cmd2, or in sequence using cmd1 && cmd2. You can also do fancy things with piping into xargs and do string manipulations using sed and awk, but I only ever copy-paste from StackOverflow for the latter if I need to use them.
You can set commands to run at startup time by adding them to your ~/.bashrc file (the tilde expands to your home directory). (They won't take effect until you restart or do source ~/.bashrc though.)
A lot of tools have CLI's (command-line interfaces) e.g. if you've pip install'd black then you can call black my_file.py to format it. One very import command-line tool is git.
Git
git is a command-line tool for software version control. In particular it enables keeping track of a fine-grained history of a software project (aka repo), one snapshot for every commit along with a commit message like 'added parser' or 'made edits', which is very useful when you've just introduced an obscure bug and need to roll back to a trusted checkpoint.

You can also easily see the diff between any pair of commits, and selectively make changes. The real magic of git is that it allows a team of collaborators to simultaneously edit a repo. GitHub (and also azure devops!) allows for storing a remote repo, to which updates are periodically pushed, and which collaborators can clone and then keep updated by pulling new code. To make simultaneous edits, collaborators typically work on different branches from the main (or master) branch, like 'dawn/improve-parsing', which are periodically merged into the main branch via a pull request after being pushed to GithHub. The initiation of a pull request is a natural point for code review. Often there's a somewhat painful step after attempting to pull or merge where the developer has to open an editor and resolve merge conflicts from simultaneous edits to the same code block that can't be automatically stacked with each other.
It's a lot of jargon (repo, clone, branch, commit, pull, push, merge, pull request (pr), as well as diff, merge conflict, not to mention add, commit message, checkout etc.). It's pretty easy to get the hang of things if you're working with people who actually use this tool every day. At its heart using git just comes down to the same handful of commands over and over again.

Here's a more in-depth guide (along with actual commands) which is pretty good https://rogerdudler.github.io/git-guide/
The main gotcha for me was when I would git add new_file.py && git commit -m 'made change' before git checkout other_branch — when you do this it looks like new_file.py has completely disappeared, but its actually still on whichever branch you were on before, and easy to get back. Conversely, if you create a new file, don't add and commit it, and then switch branches, it should still show up, so no need to worry.
Another point of confusion and frustration for me is when I would try to git pull origin main into my current local branch, and be greeted with a warning that doing so would overwrite my current changes, unless I first committed or stashed them. Well. You can indeed fix this issue by first git add'ing and commit'ing your changes. I have also several times done git stash before pulling as suggested... without realizing that I then had to git stash pop to get my changes back! Very frustrating when you have no idea what's going on and it looks like you just accidentally deleted all your code haha.
Research
Reading papers
I read 10+ papers a week for most of my time at Microsoft, tapering off towards the end. The trick is that most papers offer very incremental advances, if any e.g. a tiny change to an attention operation or positional embedding, which ultimately corresponds to like three lines of code. When I was reading papers within my more narrow domain (ML applied to source code), I would primarily hunt for a description of the data. With papers claiming some kind of architectural tweak, it's good practice to first read their graphs and tables and see if they're actually making an improvement rather than just bullshitting.

For getting wind of exciting new papers, I mostly use r/machinelearning and coworkers. I imagine you could also use twitter, conferences, and newsletters. Maybe arxiv-sanity.
In terms of getting a foundational background on transformers, I'd recommend Jay Alammar's great illustrated guide(s) https://jalammar.github.io/illustrated-transformer/. I also got a lot of value out of fast.ai's course at the start of my career as a data scientist.

Writing papers
Writing machine learning papers is fairly cookie cutter. Grab any three papers and you'll see they're all structured in nearly the same way: title, abstract, intro (including overview of main results), (optionally have a longer related works section here or at the end), the model, the data, experiments and results, the bibliography, and the appendix with miscellaneous results, implementation details, or proofs.
In addition to standard software development tools like GitHub, I've also gotten a lot of value out of jointly editing LaTeX docs on Overleaf or markdown documents in shared wikis like Roam or Notion. It can also be nice to cannibalize powerpoint presentations for the figures for the paper.
In my experience, it feels like writing the paper is more fun and about as time-consuming as the process of actually getting it submitted to a conference, which requires lots of reformatting (reminiscent of messing with font sizes and whitespace for high school essays...), extreme pettiness in the sake of "anonymity" and this boundless army of reviewer number 2's. Delenda est etc. The value drain is real and the value add is not there.

Prioritizing
Prioritizing research is hard, because you very much have to carve out your own path, and it's easy to get distracted by a hundred different experiments or questions, most of which aren't actually action relevant. One thing I like to do is have a list of planned and completed giant wins, medium wins, and small wins, where a giant win would be a great accomplishment worthy of spending an entire month, a medium win is on the scale of a week, and a small win is on the scale of half a day or so. Sometimes it will become obvious when options are laid out like that how some time sinks won't amount to much more than a tiny win, and how some giant wins are actually pretty straightforward.
Having a job
Meetings
The number of meetings I was invited to slowly ratcheted up while I was at Microsoft. I was... not a fan of most of these meetings, especially because I personally had these big switching costs (often being distracted for fifteen minutes before the meeting even started, or at least being afraid to start anything that would take longer than fifteen minutes, and often needing to recuperate after an especially boring or frustrating meeting. There was one especially embarrassing time when I silently left a meeting early (leaving myself in the Teams meeting itself while muted), and returned an hour later to see that they had been recording, and could not stop recording until everyone had technically left.
I remember my roommate Kevin, who had joined Microsoft as a program manager at the same time as me, being frustrated with how it seemed like he had to go to all these dumb meetings and after several months was still in this weird limbo where he hadn't really started his job. Finally, he had an epiphany, and exclaimed, "I've got it now! My job is meetings!" Thankfully my job is only like ten percent meetings, although it was maybe more like 25% towards the end at Microsoft.
In one moment of questionable genius I moved every meeting I could to Thursday, which started out absolutely amazing.
There were several upsides to having all my meetings scheduled for that distant Thursday, chief among them that I had this beautiful defragmented calendar, and that I could selectively screenshot it to make myself appear to be in very high demand. As it turned out, I don't know, I feel like I was less invested in my job the rest of the week, and I kind of ended up having the same conversation several times in a row, which was of little value. The biggest shortcoming was that, well, a lot of my meetings were one-on-ones, and I actually really liked most of those meetings, so ideally I would have spaced those out and collapsed the more boring meetings where I wouldn't actually participate.

Anthropic actually has a pretty similar strategy in that recurring meetings are only allowed M/W/F, and a premium is placed on long uninterrupted blocks. Notably, we don't consider pair programming to fall in the meeting category, and are very liberal about pairing up throughout the week. Pair programming is extremely helpful in my experience when one person is much more knowledgeable about some poorly documented or hard-to-debug workflow than the noobier person, or when the two people have complementary knowledge (e.g. if a coworker is trying to add spark or async support to some code I wrote, and I'm inexperienced with those tools.) It's not super useful when there's something routine or completely new that someone just has to bang their against for an hour or two.
Presentations
I gave maybe ten powerpoint presentations at Microsoft, on my own work and on papers that seemed important, and I was invited to many more. I strove to make it very easy for someone to get the brunt of my presentation without having to sit through the whole thing, mostly by sharing the slides ahead of time and starting with a quick high-level overview. I also tried to make a point of sharing lots of concrete examples (it can be very easy to succumb to the curse of knowledge and only talk about your work in the abstract), and to interleave opportunities for questions throughout (it's kind of lame if someone gets lost at the very beginning, and having ongoing Q&A's will make people much more engaged.) I also liked giving live demos (especially in low-stakes meetings). For the highest stakes presentations, e.g. to CEO Satya himself, we would record things ahead of time.
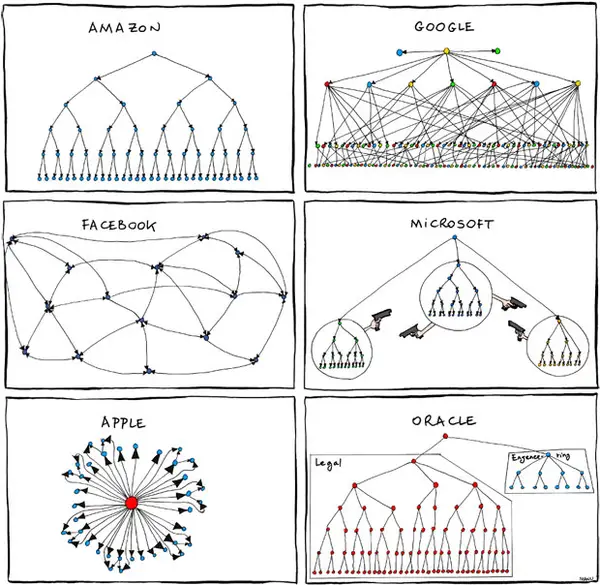
Politics
The main unsavory way I saw politics play out at Microsoft is that I felt like a lot of my collaborations were really weird, and it wasn't super clear if the value add was helping each other or appearing to help each other. I'm not going to give any details here since it seems inappropriate, but I will share this comic about inter-team dynamics from the Steve Ballmer days:
Investing
One of the best parts of having a job is getting paid. On brilliant advice from the man who arguably went from mere millions to decabillions faster than anyone in modern history, I've put a large chunk of my money in leveraged index funds and etfs. Which, yeah, have been doing quite well (see SPXL 30x'ing since its inception thirteen years ago in late 2008), during which time SPY has merely quintupled). Not nearly as meteoric a rise as FTT, but still pretty good.
Data Science and the dev cycle
I learned a few good habits from cloning cookiecutter-data-science, which automatically gives you a readme.md, a setup.py, separate source folders for data work and modeling work, a notebooks folder which is split into messy protoyping and polished sharing, and separated data folders for raw, interim, and process data. (There are also several extraneous files admittedly.)
https://github.com/drivendata/cookiecutter-data-science
An important maxim is to make sure to look at the data early and often! That will give you a sense of how hard the problem is and how good your model is, and it will make it obvious when bugs like off-by-one errors or bad shuffling inevitably creep in. Related, it's extremely helpful to monitor your training runs using visualizations from wandb or tensorboard.
One thing that I've been hit over the head with time and time again is that every coding project takes ten times as long as I think it should. I have termed this phenomenon Dawn's rule, that is: Everything takes ten times longer than it should, even after accounting for Dawn's rule (I believe its less quantitative but more canonical name would be https://en.wikipedia.org/wiki/Hofstadter's_law). It has been a remarkably robust phenomenon in my experience! I think there are a couple reasons for this.
First, I sometimes conflate programming time with clock time. Indeed, it may only take four hours to do a certain project, but those hours could be very spread out, especially if I deprioritize the project before I'm finished. The minimum serial time might even be fairly long if I need to wait for a run to finish.
Second, because there is a long tail in terms of how long any given step of a project will take, and it's very easy to get stuck hard on a bug that I hadn't even considered in the first place. Erik Bern argues that programmers are actually pretty good at estimating the median time it will take to complete a step, but are very bad at combining those projections to estimate the average time a project will take https://erikbern.com/2019/04/15/why-software-projects-take-longer-than-you-think-a-statistical-model.html
Finally there is often scope creep between the quick plan in my head and what I actually want to accomplish in practice. That is, I might be imagining how long it will take to get a hold of the data and train a baseline model, but really there are many more steps in the data science process.
Using an IDE
Despite working on the developer tools team trying to improve VS Code extensions for nearly two years, I didn't actually start using an IDE until I joined Anthropic and started using pycharm (I had used VS Code for a couple weeks at Microsoft, but gave it up after this one update messed with my port forwarding. Ironically I found the intelligent autocompletion more annoying than simply doing dir(my_var) and getting an exhaustive lists of options, and I was working with a small codebase (mostly my own code) where I didn't benefit from goto. For nearly two years I used jupyter as my main editor of all things.) Well! Now I do use an actual IDE, pycharm, and it has several great features, in addition to the basic syntax highlighting provided by jupyter. The most mind blowing are goto (go to a function or variable's definition) and autocomplete. I also like warnings from pylint, and I have these hooks for autoblack (very popular python formatter) and auto-isort (groups imports by which library they come from and sorts them alphabetically) on file save. Anthropic also insists on autoflake (removes unused imports, and can also remove unused variables) before merging. IT TURNS OUT that it's possible to write a code snippet that is not black even after doing black and then isort — this gave me a very hard time for several weeks before I realized what was going on.
A data scientist should also know how to use vim (or emacs), since there are inevitably times when you have to edit a file on the command line. I've found the built-in tutorial vimtutor to be very helpful :). It clarified visual vs. insert vs. normal mode for me, how to find and replace, delete a line, highlight text and copy and paste etc. It's brilliantly interactive.

Gear (loosely construed)
I spend about eight hours a day wearing these godly noise-canceling headphones from Sony, which just two years ago cost $300 and now are as cheap as $100 on ebay https://www.ebay.com/p/17023331317?thm=2000. The one day I forgot to bring them to work, I was so overwhelmed by all the office noise that I grabbed a stack of papers and spent the entire day camped out in a different room. I keep an extra pair downstairs in addition to the one in my home office, just so that I can don them on short notice if I want to work on a problem or tune someone out. I also have spotify premium to complement my headphones.
I spent most of my adolescence addicted to various computer games and websites, which I will refrain from naming here out of respect for info hazards. I FINALLY solved this lifelong problem my first year at Microsoft by paying a token $50 or so for the program FocusMe, which does a very robust job of blocking any site I tell it to. Another premium option is RescueTime, which additionally provides excellent fine-grained time-tracking (essential for doing any kind of life-hacking experiments, although Apple's default Screen Time is already pretty good). There are also free browser extensions like leechblock which do a similar job, albeit with more trivial workarounds. I also have special tricks for important websites that I still want to access, but simply in a less addicting way. Specifically I have FaceBook Newsfeed Eradicator, and I use ublock origin to block the comments and video recommendations on YouTube, see https://github.com/JesseDrain/Less-Addictive-YouTube. Another small trick for avoiding distraction is to keep my phone out of sight, literally a foot away, but buried under a sweater or pillow so that it's effectively invisible.

I have this awesome monitor 43" (tv) 4k monitor which I got from amazon for $300 https://www.amazon.com/TCL-43S425-Inch-Ultra-Smart/dp/B07DK5PZFY, a steal compared to the price of a comparable monitor marketed specifically for computers rather than as a tv. I turned it vertically (resting it against the wall...) and can fit over 300 vertical lines of code :), and I can comfortably fit 600 LOC if I split screen. Remarkably it only weights fifteen pounds. The main downside is that it won't fit in a suitcase and is expensive to ship. Also I can get kind of enraged when reduced to 20 LOC on my laptop...
I also have these extremely bright 60W cornbulb LED lights, which give my third-floor office a kind of mad scientist vibe when viewed from the street. They're especially good at combating 5pm winter sunsets. I also have a personal fan, which helps with those 110 degree Seattle afternoons we apparently get now oh my god global warming is real, as well as an air purifier for when the world is on fire.
Stimulants like caffeine, adderall, and modafinil are magic. If I could always be on stimulants without developing tolerance I... would be like 2-3x as productive and 20% happier. It's this one dollar pill and then you get like an extra five hours of highly engrossing work. People do stay on adderall and modafinil indefinitely while at least reporting only partial or no tolerance (see section four here https://slatestarcodex.com/2017/12/28/adderall-risks-much-more-than-you-wanted-to-know/, and the penultimate section https://slatestarcodex.com/2014/02/16/nootropics-survey-results-and-analysis/ (personally I feel like people develop tolerance to caffeine very quickly than to adderall and modafinil)). But, well, I still want to have some more long-term tracking data from RescueTime and Screen Time before I personally start taking these drugs on anything like a daily basis.

Shortcuts
I'm not talking about shortcuts like taking drugs to get ahead, I mean keyboard shortcuts. The keyboard shortcuts that I use the most are:
- cmd+c, cmd+v (copy and paste) (and cmd+x for cut)
- windows also has a clipboard history you can access with windows+v, but sadly you need a paid program like Alfred to get a similar tool for mac
- cmd+z (undo) (and cmd+y for redo)
- cmd+f (find)
- cmd+s (save)
- cmd+shift+4 (screenshot) (add ctrl to save to clipboard!)
- cmd+arrow-key (jump to start or end of line) (+shift to highlight
- cmd+t (open new tab) (cmd+shift+t to restore a tab you just closed)
- cmd+/ (comment out highlighted code)
- fn+backspace (delete)
- cmd+l (go to search bar)
- cmd+ctrl+q (lock screen)
- cmd+shift+f (full screen)
- cmd+a (highlight all)
- cmd+w (close tab)
- cmd+, (see preferences)
- cmd+n (new window) (cmd+shift+p to make it a private window)
- cmd+r (refresh page)
- cmd+k (add hyperlink)
If you download a tool like spectacle or rectangle, you can also use shortcuts to move windows around, which makes it very easy to split screen.
The trackpad shortcuts I use most often are:
- use two fingers to scroll up and down
- use two fingers to go back or forward a page
- use four fingers to switch between desktops (essential for using a single monitor imo)
My favorite mouse shortcuts are:
- Double click to highlight the current word
- Triple click to highlight the current line
- cmd+click to open link in new tab
Another kind of shortcut is snippets. On a coworker's suggestion I bought Alfred, this mac productivity suite. One cool feature is it lets you replace a specified command with a saved snippet. For instance, I have a snippet !reload which Alfred turns into
%load_ext autoreload
%autoreload 2Convenient :), and kind of magical.